Researchers build a Wikipedia for resistant bacteria
Date: 26.10.2022
In the future, even a small infection can become life-threatening for people if disease-causing bacteria become resistant to traditional treatment with antibiotics.
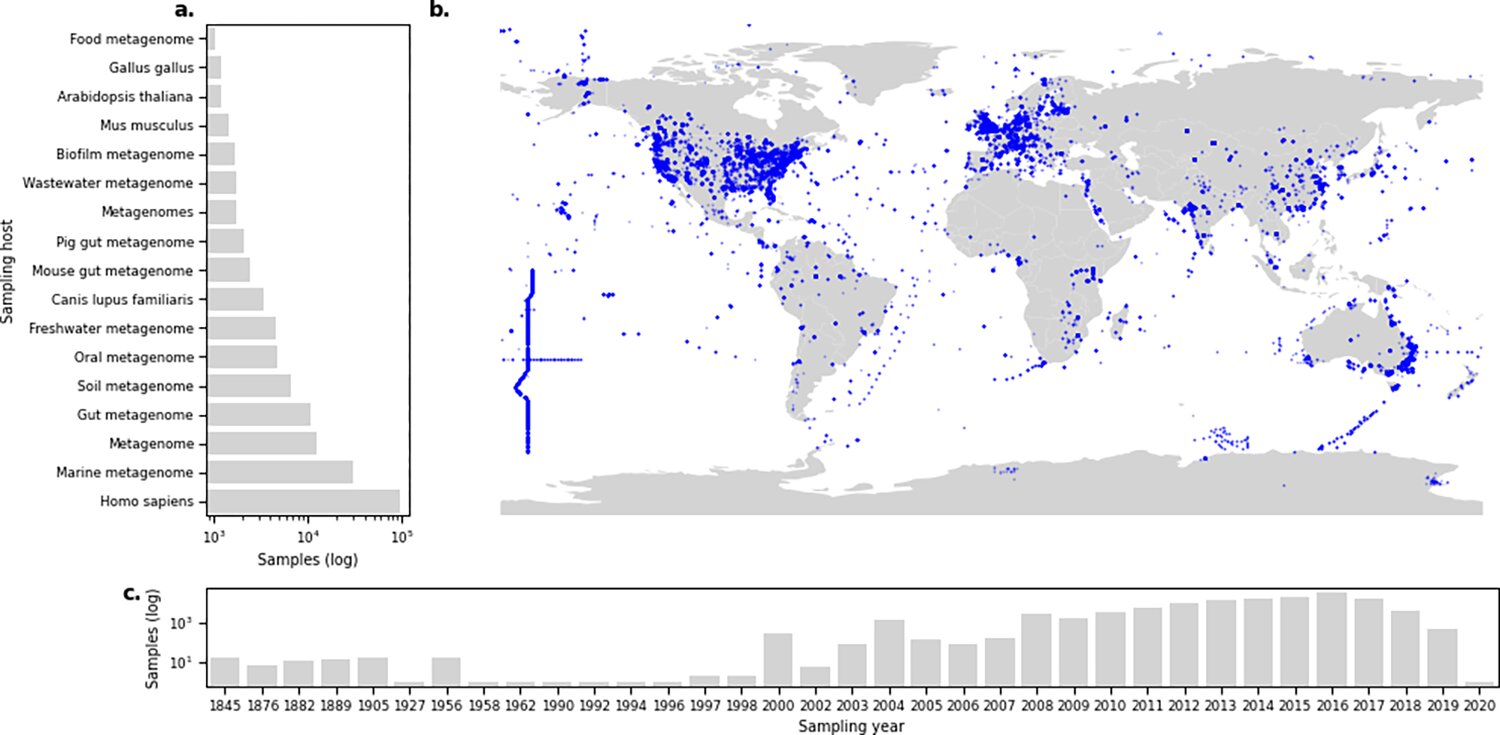 Based on 214,000 microbiome samples, DTU researchers have created a freely accessible platform that shows where in the world different types of resistant bacteria are found and in what quantities.
Based on 214,000 microbiome samples, DTU researchers have created a freely accessible platform that shows where in the world different types of resistant bacteria are found and in what quantities.
To gain an understanding of how antibiotic resistance is spreading across the world, it is important to know where, which and how many resistance genes are found in all the environments that surround us. The genes that provide resistance can spread between animals, humans, and the environment.
Researchers from the DTU National Food Institute have analyzed 214,000 samples from, among other things, animals, humans, and soil, and organized them in a way that makes it possible for others to use them. The goal is to create a catalog of resistant bacteria that spans countries, people, and environments.
"You can compare it to a large encyclopedia, like Wikipedia, which collects knowledge from many different sources and organizes it in a way so that everyone can access it. In the same way, we collect data on antibiotic resistance in bacteria and share it with everyone," she says. The 214,000 samples together take up almost 300 terabytes and it takes months to analyze it on a high-performance-computer.
Image source: Martiny et al. (2022), PLoS Biology.























