Supercomputers simulate new pathways for potential RNA virus treatment
Date: 6.1.2021
University of New Hampshire (UNH) researchers recently used Comet at the San Diego Supercomputer Center at UC San Diego and Stampede2 at the Texas Advanced Computing Center to identify new inhibitor binding/unbinding pathways in an RNA-based virus.
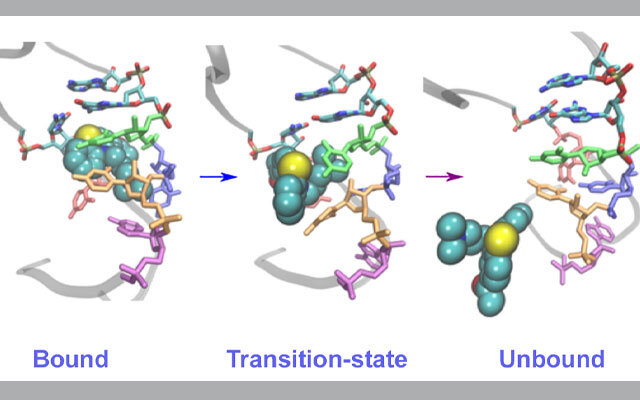 The findings could be beneficial in understanding how these inhibitors react and potentially help develop a new generation of drugs to target viruses with high death rates, such as HIV-1, Zika, Ebola, and SARS-CoV2, the virus that causes COVID-19.
The findings could be beneficial in understanding how these inhibitors react and potentially help develop a new generation of drugs to target viruses with high death rates, such as HIV-1, Zika, Ebola, and SARS-CoV2, the virus that causes COVID-19.
"When we first started this research, we never anticipated that we'd be in the midst of a pandemic caused by an RNA virus," said Harish Vashisth, associate professor of chemical engineering at UNH. "As these types of viruses emerge, our findings will hopefully offer an enhanced understanding of how viral RNAs interact with inhibitors and be used to design better treatments."
Vashisth and his team created molecular dynamics simulations using the Comet and Stampede2 supercomputers to look specifically at an RNA fragment from the HIV-1 virus and its interaction with acetylpromazine, a small molecule that is known to interfere with the virus replication process.























